Completing my extension of geiger into object oriented design whereby I can easily apply the Monte Carlo approach to test fits using function calls to model fits such as update() and simulate(). A quick convenient example: AIC would prefer (though not by munch) the delta transform model over the Brownian motion model; a conclusion not supported by the Monte Carlo test, which suggests there is not enough power to choose between these models:
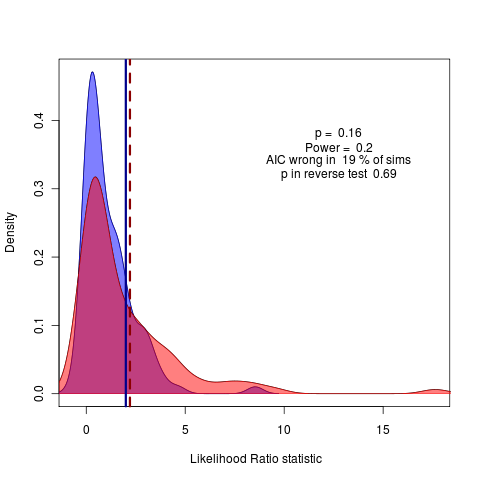
The density kernel estimation does make it appear that some values are smaller than zero though. Recall all estimates are from the replicates directly, so this doesn’t introduce any errors in the numbers reported, only the visualization; still it is annoying. Also note this is just with 100 replicates, not 2000, since I wanted a quick example and geiger is much slower than ouch.
Will see if I can generate enough interesting examples out of the Anoles data or if it would be good to include some simulated data (and maybe some simulated trees with different structures). Simulating under OU, fitting lambda and bm models. Alpha = 1 and Alpha = 2
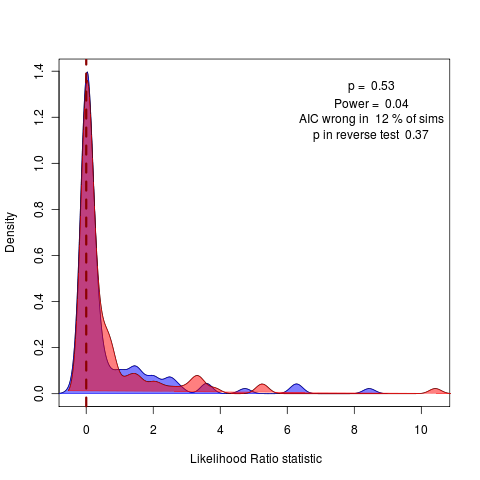
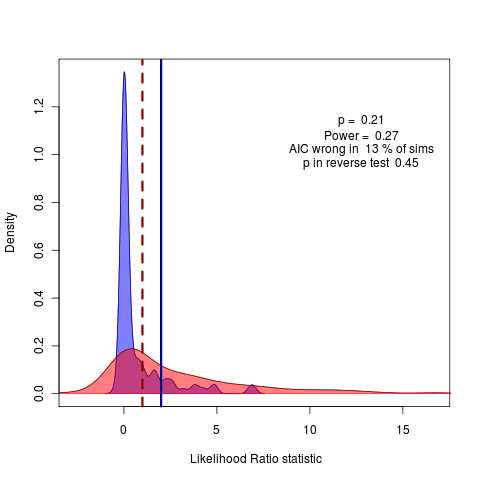
(code)
Ah, the strange likelihood ratios under the test simulation are possibly the result of a bug in the simulation script, need to rescale tree to estimate the phylogenetic mean. Confirmed this works with ape’s rTraitCont simulation which can simulate BM (and transformed tree versions of course) and OU) See code changes. Still, surprisingly wide distribution of likelihoods under simulations of the lambda model.
Consider the Geiger geospiza data for example, which shows the same wide uncertainty in simulating (n=2000 now) under the test model:
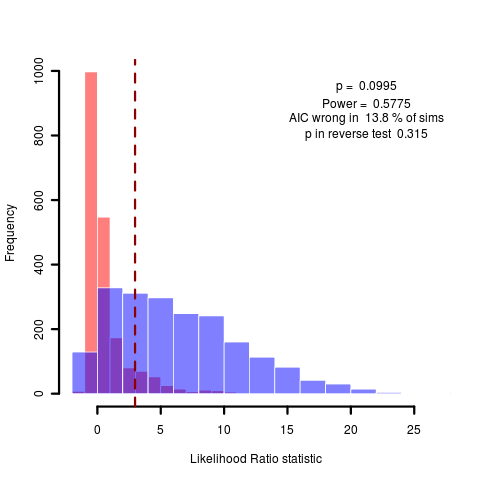
Beginning to wonder if it’s worth bothering to explain how this works in the context of type I and type II error, vs just measuring the divergence between the two distributions. The important thing is realizing that its the likelihood ratio that it’s key to look at, and to look at under both distributions. a rather subtle thing.
Also looks like it may be worth porting tree transforms into OUCH just for the purposes of doing this analysis at higher speed for the fits. Should profile the code first though to make sure.