Runs: Mon midnight:
parrotfish.R with updated SANN parameters and corrected output parameter labeling (always {Xo, alpha, sigma, theta} ordering), implemented on zero, nice 19. General model (“wright”) still not outperforming others, so still un-converged.
[flickr]5808279169/[/flickr]
MCMC runs of gape.y and close on one, at nice 19 and 18 1e5 reps. (Alex running matlab nice 10). protrusion.y at nice 11, 1e6 reps.
open at nice 18 on zero, 1e6 reps.
Runs: Tues Morning
8am: parrotfish.R 19 on one done, rest running. added gape.y at 5e4 on zero at n18. done.
open, close on zero, 1e5, 5e4. done.
11am: open on zero, n17, 1e6 gen, and close on zero, n19, 1e6 gen.
Summary Results
(Click on image for parameter details, hashtag to code in git).
Open Lever arm, 1e6 gens
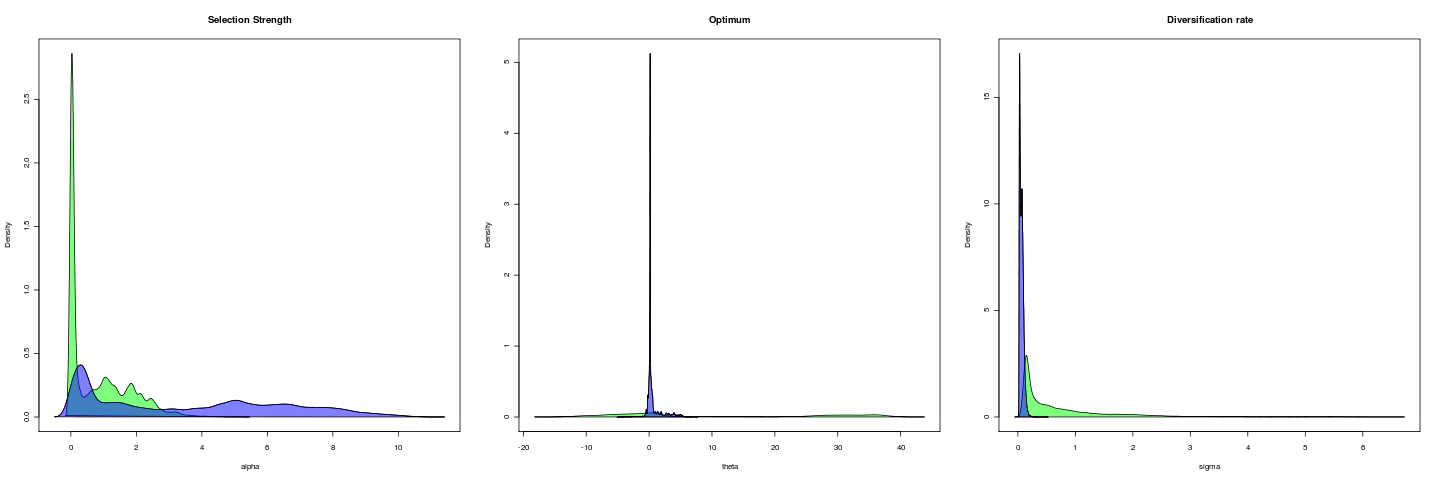
general_mcmc(data = labrid[open], tree = labrid, regimes = intramandibular, alpha = 0.1, sigma = 0.1, MaxTime = 1e+06, indep = 100)Despite the 1e6 gen probably far from stationary still: 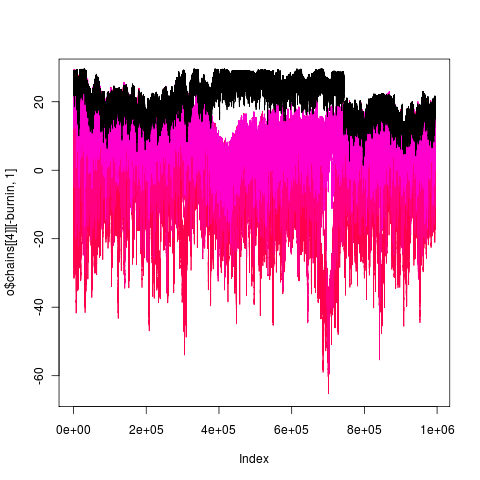
Compare to:
Opening lever arm, 1e5 gen
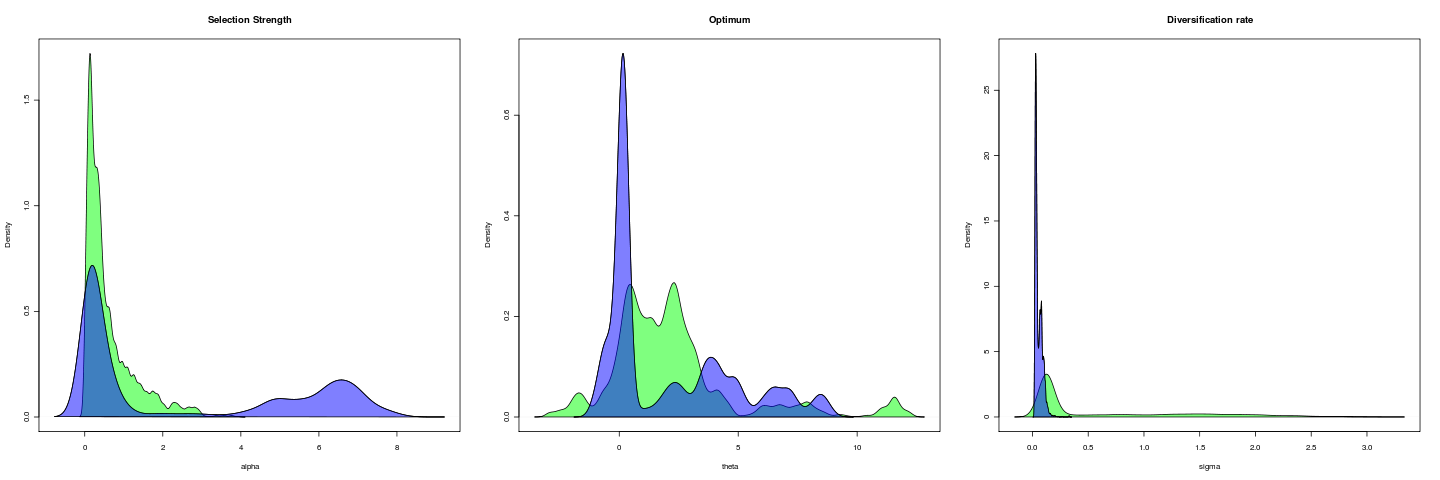
general_mcmc(data = labrid[open], tree = labrid, regimes = intramandibular, alpha = 0.1, sigma = 0.1, MaxTime = 1e+05, indep = 100)
Size-corrected gape, 5e5 gen
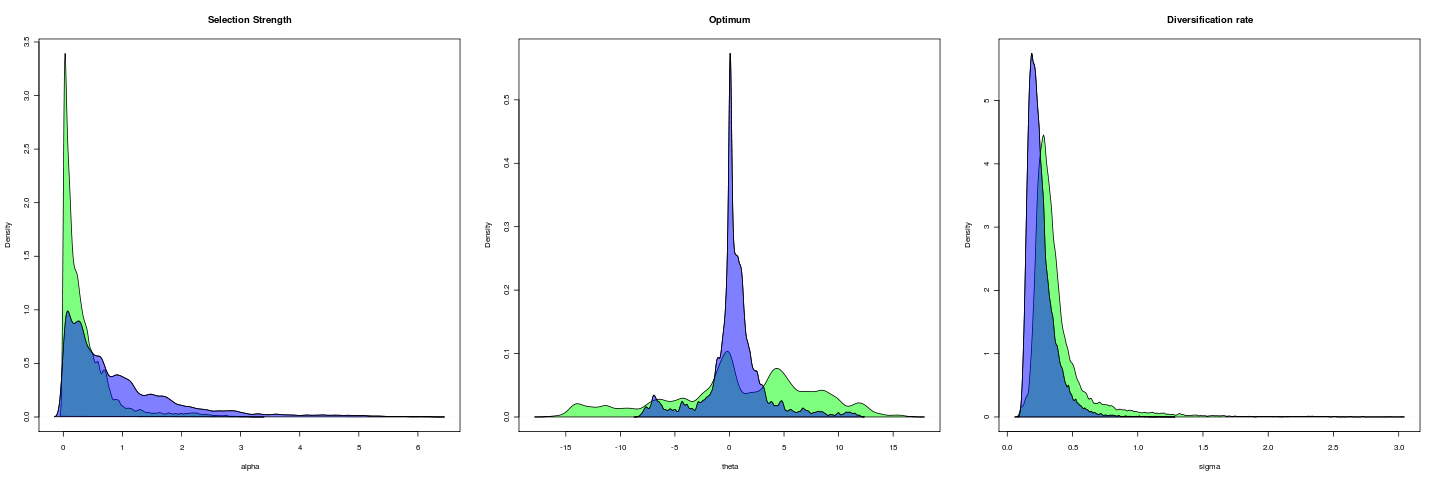
Closing lever ratio, 1e5 gen
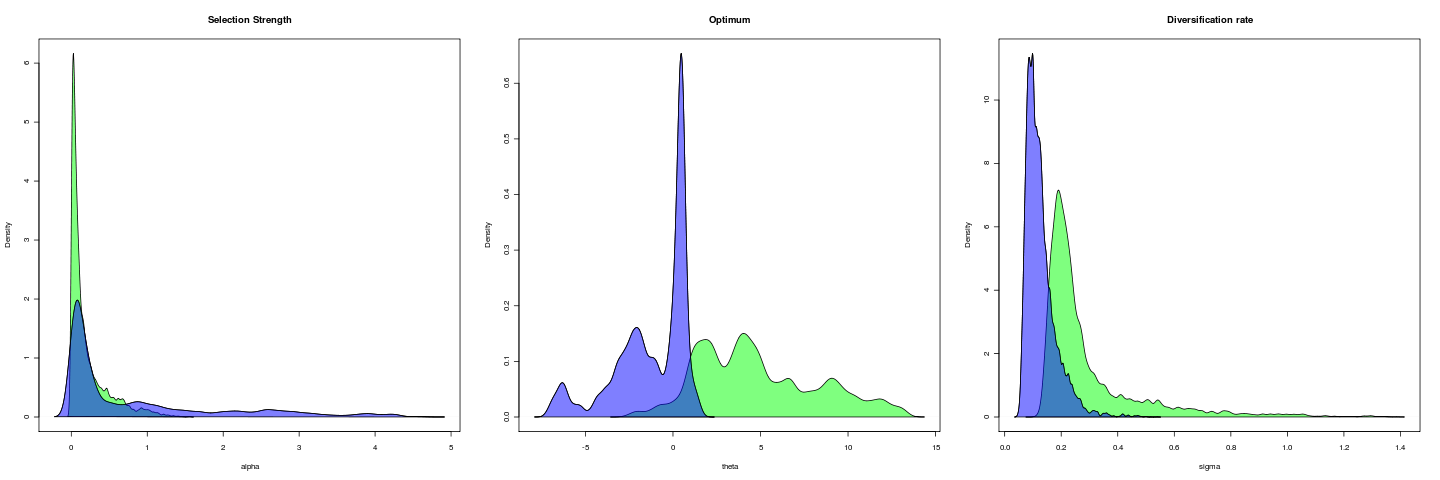
All runs today: [flickr-gallery mode=“search” tags=“phylogenetics” min_upload_date=“2011-06-07 00:00:00” max_upload_date=“2011-06-08 9:59:59”]
Afternoon: Code Updates
Rewrote the model with closures and generics. Allows model to be specified by identifying which parameters are global, which are local (and if alpha is fixed). Other combinations (global sigma, indep alpha and theta) may also be interesting. Keeps code completely functionalized, with single model (multiTypeOU) for maximum likelihood fit of any model, and single type to MCMCMC any model (phylo_mcmc, probably needs a better name).
In Progress / To Do
Testing convergence and power against simulated datasets.
Error handling for the simulation method–why does this sometimes give NAs? Eee gads output mean can be variable.
Unit test cases for all functions!
Remove deprecated separate function calls for wright, brownie, etc, and update the code examples that use them (i.e. loop_over_models_traits_regimes.R and all the parrotfish examples)
Practice Presentation
Lab meeting was moved to earlier in the day, which I missed. Hopefully will reschedule. See notes on practice presentation.
Databases projects
PLOS:
Tested out functions Scott has so far in the PLOS API, sent feedback.
Some test runs of queries, troubleshooting.
See details on the project issues tracker page