Bootstrapping option
Needs a method to pass the optional optim arguments to the update function, such that update uses the same algorithm as the initial fit. Required being a little clever:
[gist id=“1010999”]
Done. but perhaps we won’t always want to bootstrap under this? I suppose it is good practice but slow! Shoot, now I need a lot of really large computers really soon.
Excellent suggestion for better ways to do this from statscicomp list. match.call looks very promising as it avoids making a copy, but risks being evaluated in the wrong environment, which my solution above avoids.
# Using match.call()
#--------------------
f = function(y, ... ) {
myCall = match.call()
list(myCall = myCall, ans = y + dnorm(...))
<div>}
a = f(1, x = 4, sd = 2, mean = 4)> length(a$myCall)</div>
[1] 5
> a$myCall
f(y = 1, x = 4, sd = 2, mean = 4)
> a$myCall[[1]] # the function
f
> a$myCall[[2]] # represents the first argument, y
[1] 1
# Now I want to change this second value (representing y) to a 2
> a$myCall[[2]] = 2
> a$myCall
f(y = 2, x = 4, sd = 2, mean = 4)
eval(a$myCall)
$myCall
f(y = 2, x = 4, sd = 2, mean = 4)
$ans
[1] 2.199471
# Using closures
#---------------
set_up_f = function(...) {
fFn = function(y) y + dnorm(...)
}
f = set_up_f(x = 4, sd = 2, mean = 4)
f(1)
# 1.199471
f(2)
# 2.199471
- Note: parallelization function can use pre-initialized parallelization, but should warn that it is using this setup and report how many processors are being used vs requested.
General against Brownie, General against Ouch:
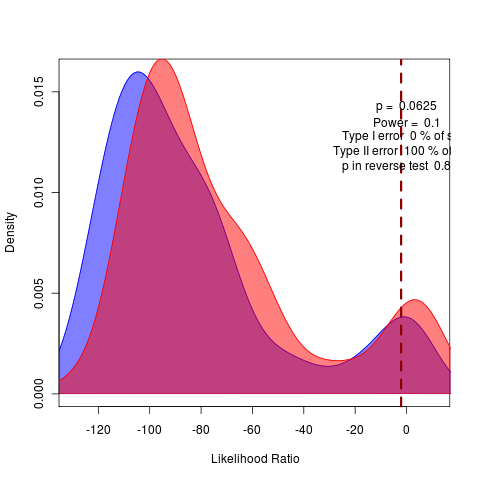
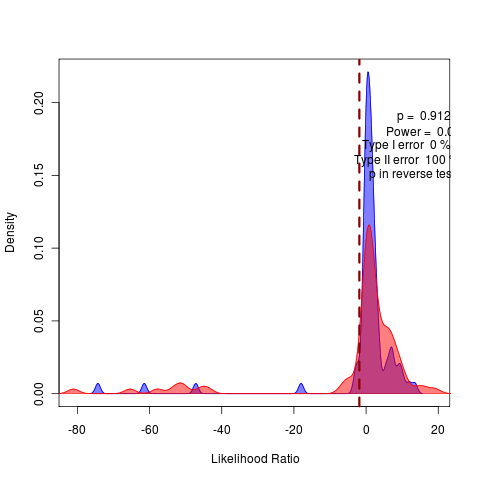
rerunning nice 17 the long SANN to do model comparison of likelihoods before attempting bootstrapping.
MCMCMC option
Opening example
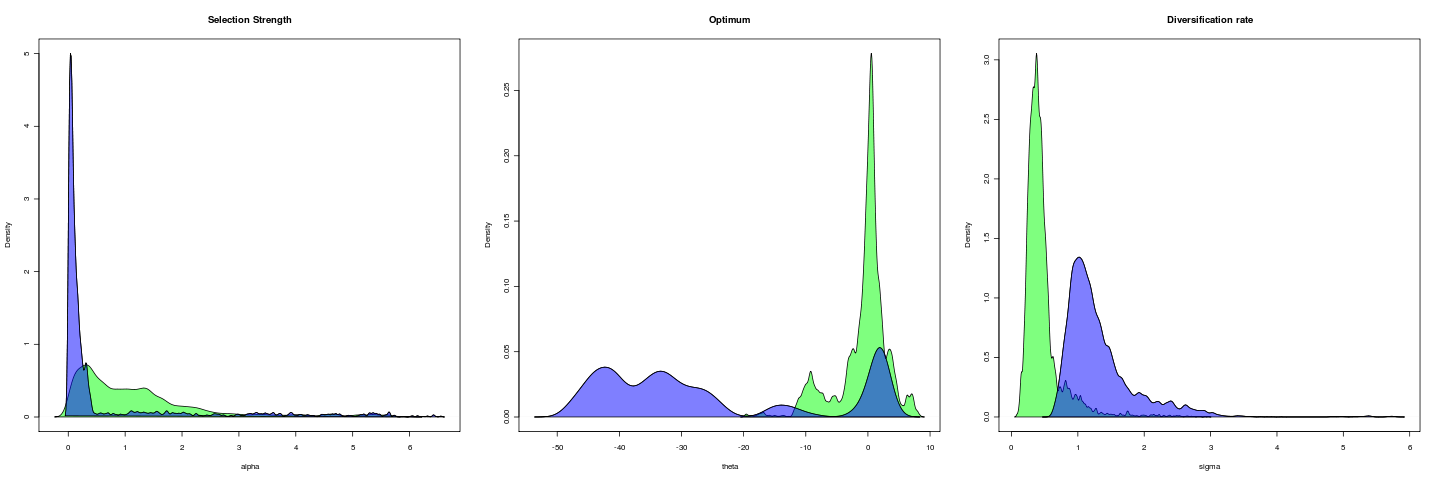
Trait is untransformed gape; gape.x.
Mixing of this model (1e5 generations), more transparent is hotter:
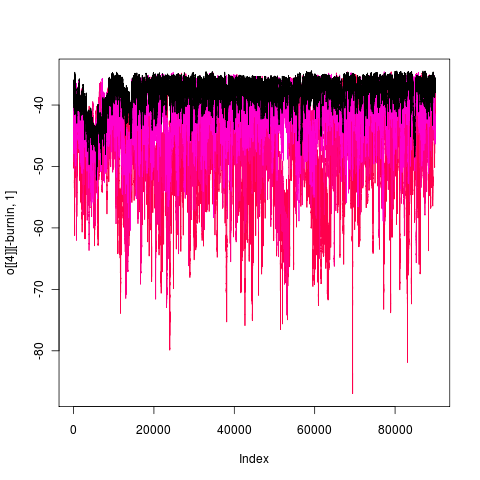
gape.y running 1e6 generations, nice 18
open (opening lever ratio running 1e4 gens, nice 16)
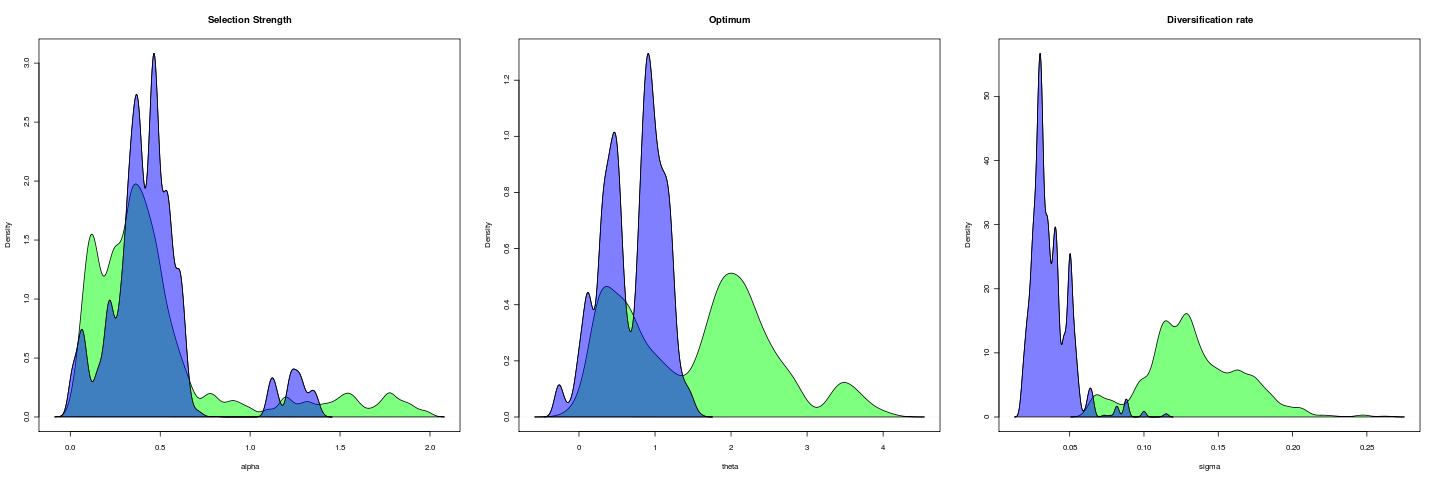
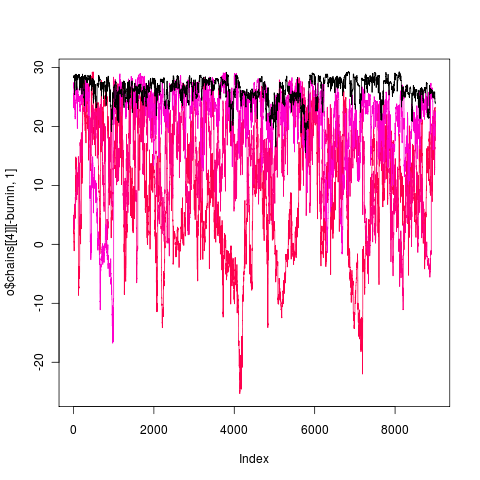
rerunning 1e5 gens, nice 14
Other stuff
Algorithms group meeting on EM today, to be continued.
Working on evolution presentation, see draft notes.
Some work on RR workflow tools, see draft notes until posted.
Registered for CSGF conference and HPC workshop
Julie’s questions on likelihood of SIR-type model simulation (in email)
To Do
Power in phylogenies manuscript finalize!
Assemble MCMC and bootstrapping results, write remaining Evolution practice talk
Hastings paper