- Fixed bug causing montecarlotest() calls not to complete: in likelihood_bifurcation_models, LTC error checking uses any() to see if any Vx are negative, returns NA if none are negative but has NAs. rm.na=TRUE fixed this.
- GetParNames works, probably worth an appendix section to discuss parameter bootstraps as well, particularly in the cases of low power. Updated codes to turn this flag back on and am re-running.
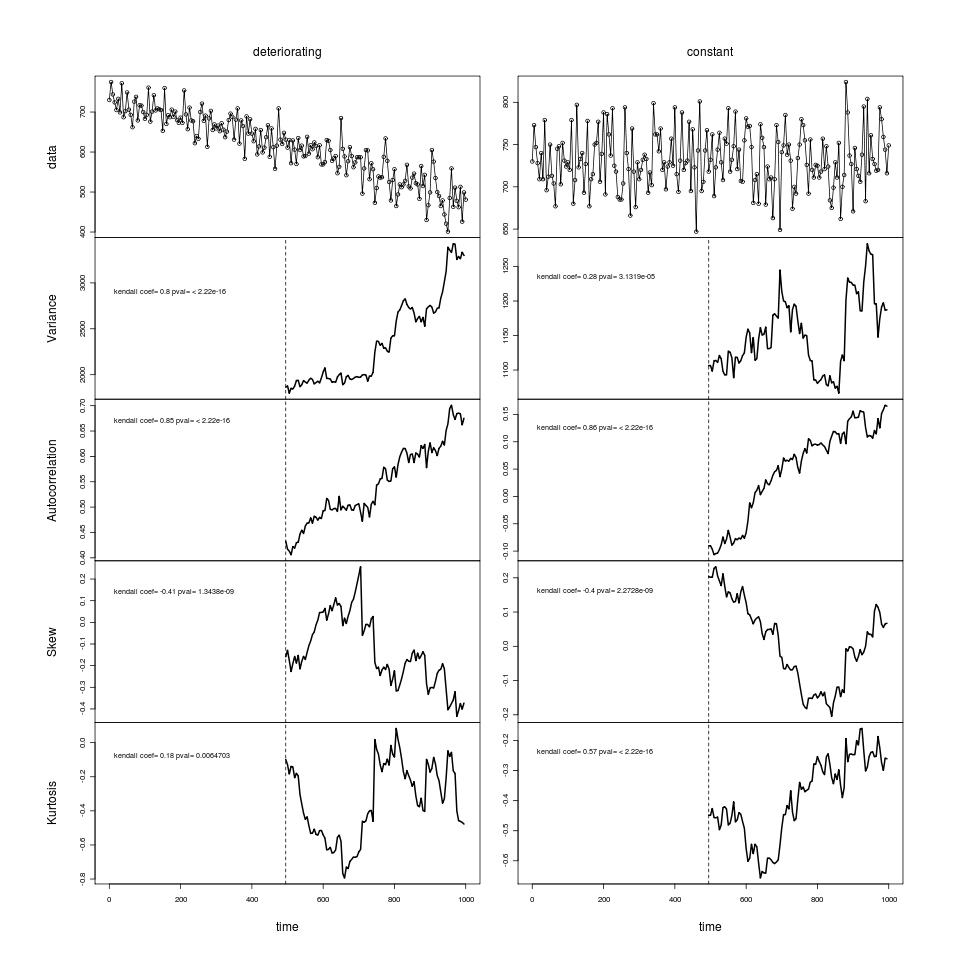
- Figure3 plot has been using the 160 bootstrap replicates, updated to the 2000 replicates. (replaces data id=35555677786 with id=35563325713). Not sure why data write added the “3” to the id code in front of the traditional flickrid.
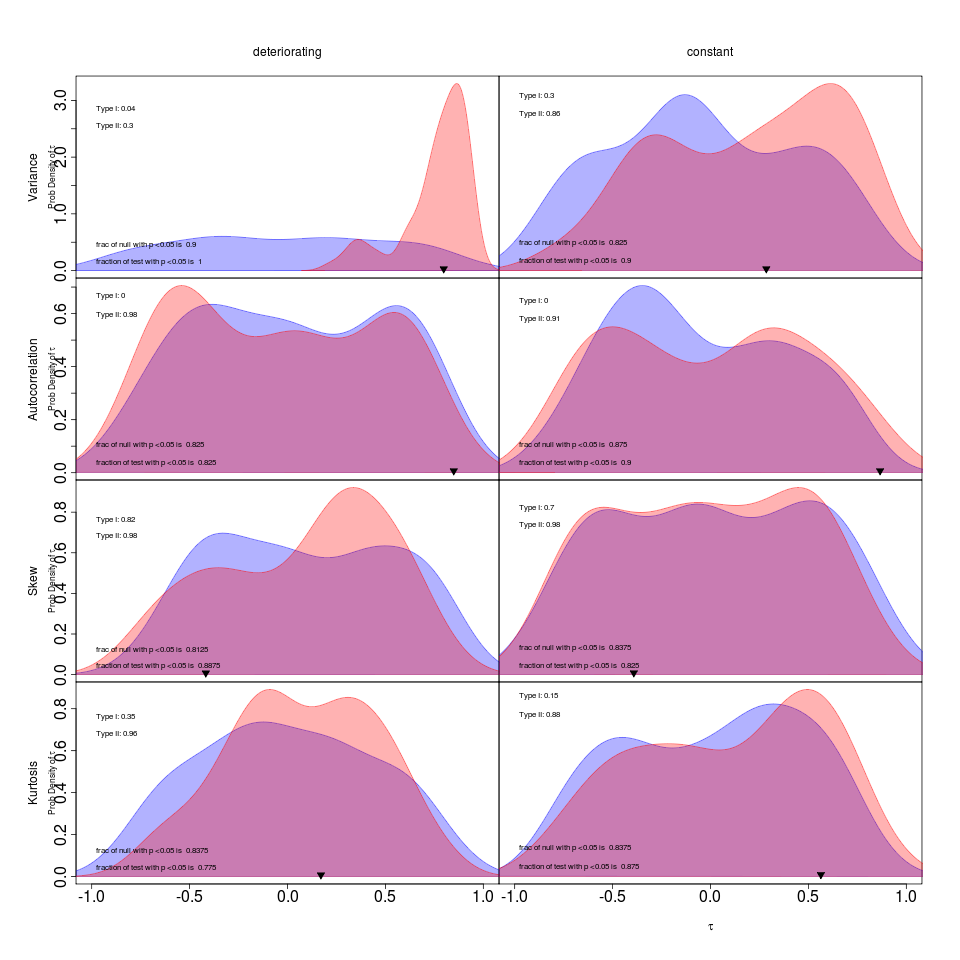
- Writing AIC section. Made plots of relative error rates. Updates to AIC plot controls to include percent error in the legend rather than as extra text. Curious if I should show both False alarm and missed event rates, maybe confusing how the test and null distributions relate to models estimated from the same dataset. appendix_modelchoice.R
- Started writing a parameter bootstrap section. Will see if we choose to keep this. Will have to wait on runs now running on zero, nice 19 & 18, deut_likelihood.R and ibm_likelihood.R
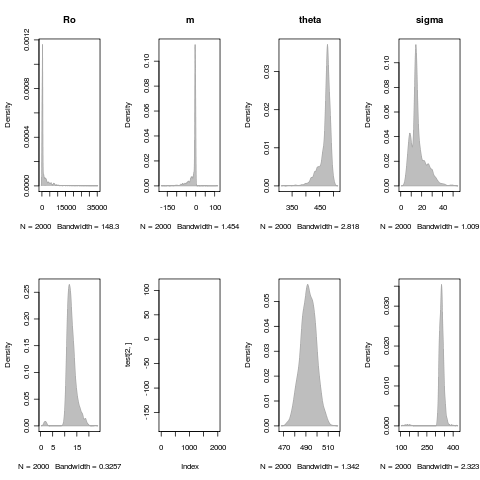
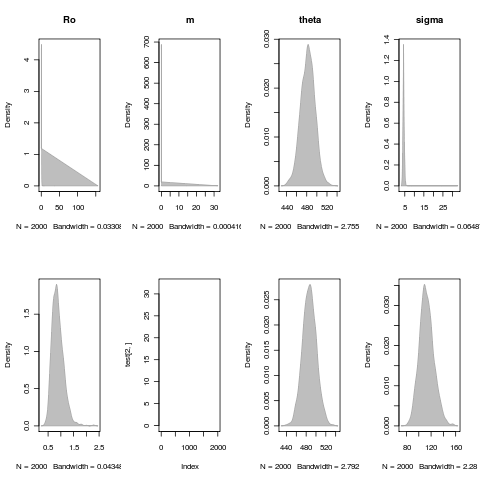
Exploring parameter distributions
Need some more exploration, remove any cases that have failed convergence, etc. All are for the LSN model fit: (parameter_dist.R)
Deteriorating environment fit
Constant Env Fit
Deuterium, Glaciation III
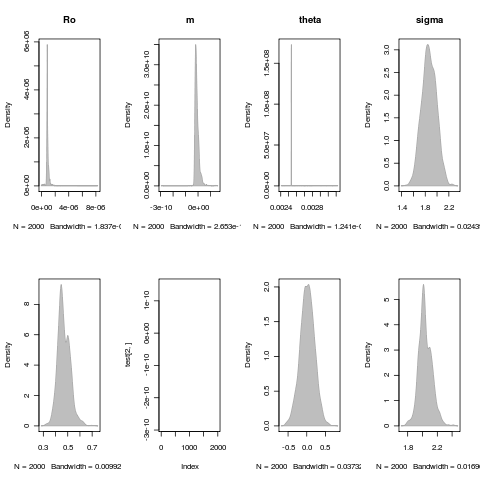
- Just added a convergence indicator to parameter list. not included on above runs. Rerunning figure3.R code on one, nice 19 to reflect these updates.
- Running 80 replicates on figure3.R at nice 1 on zero for exploration of convergence. Should consider convergence issues directly in montecarlotest().
- Running model_choice.R in a way that saves the plotting objects, need to then add plots to appendix.R
IBM Results