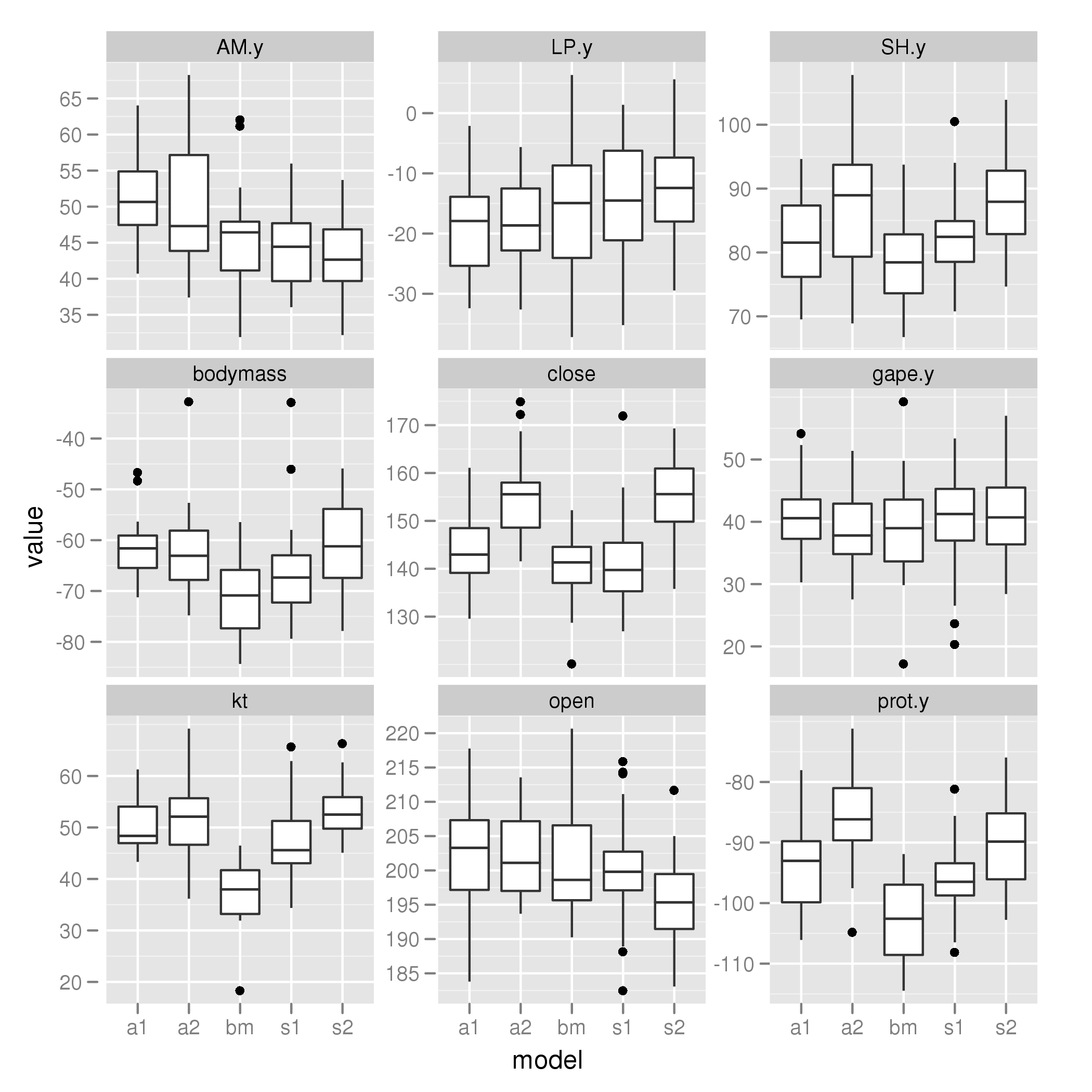
Two shifts
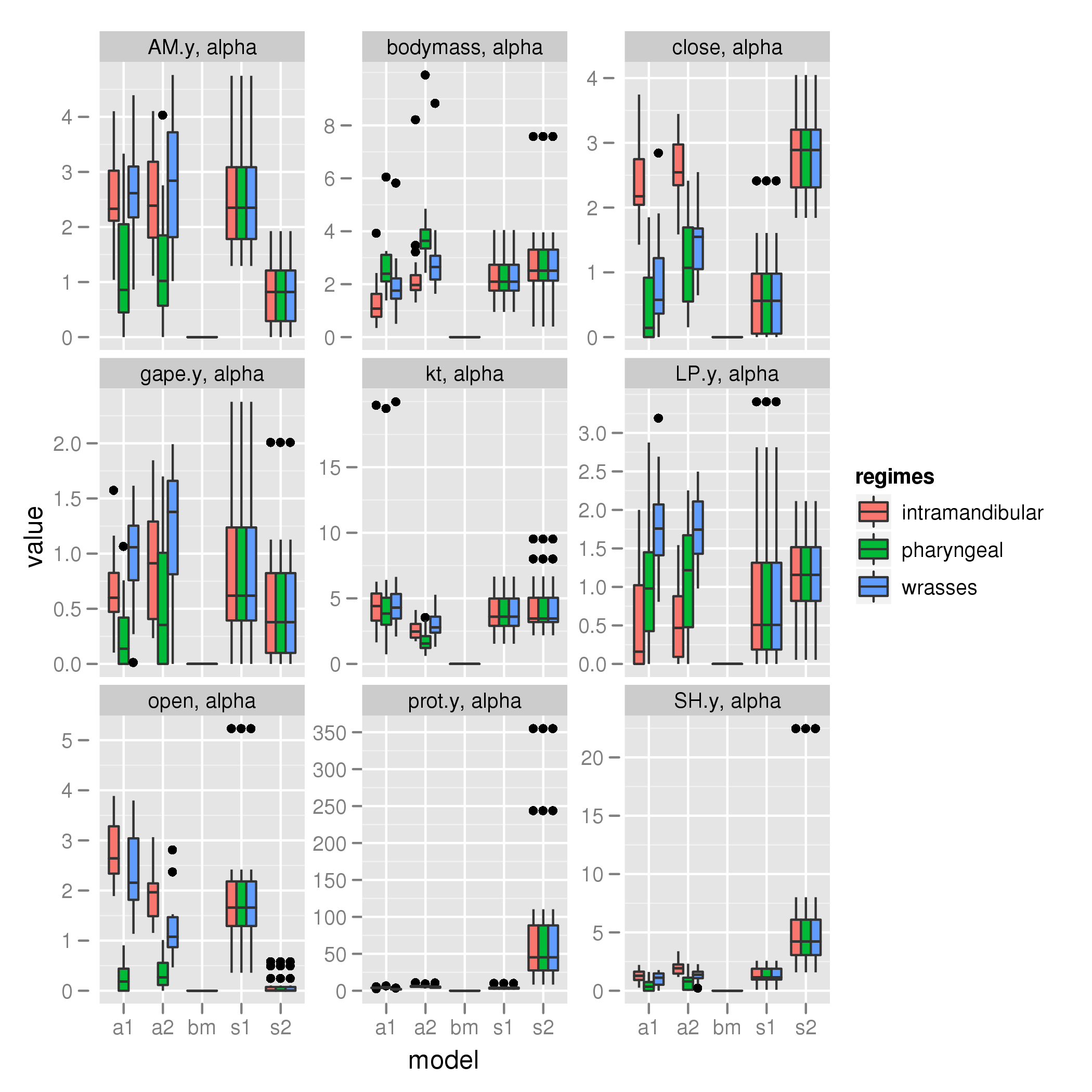
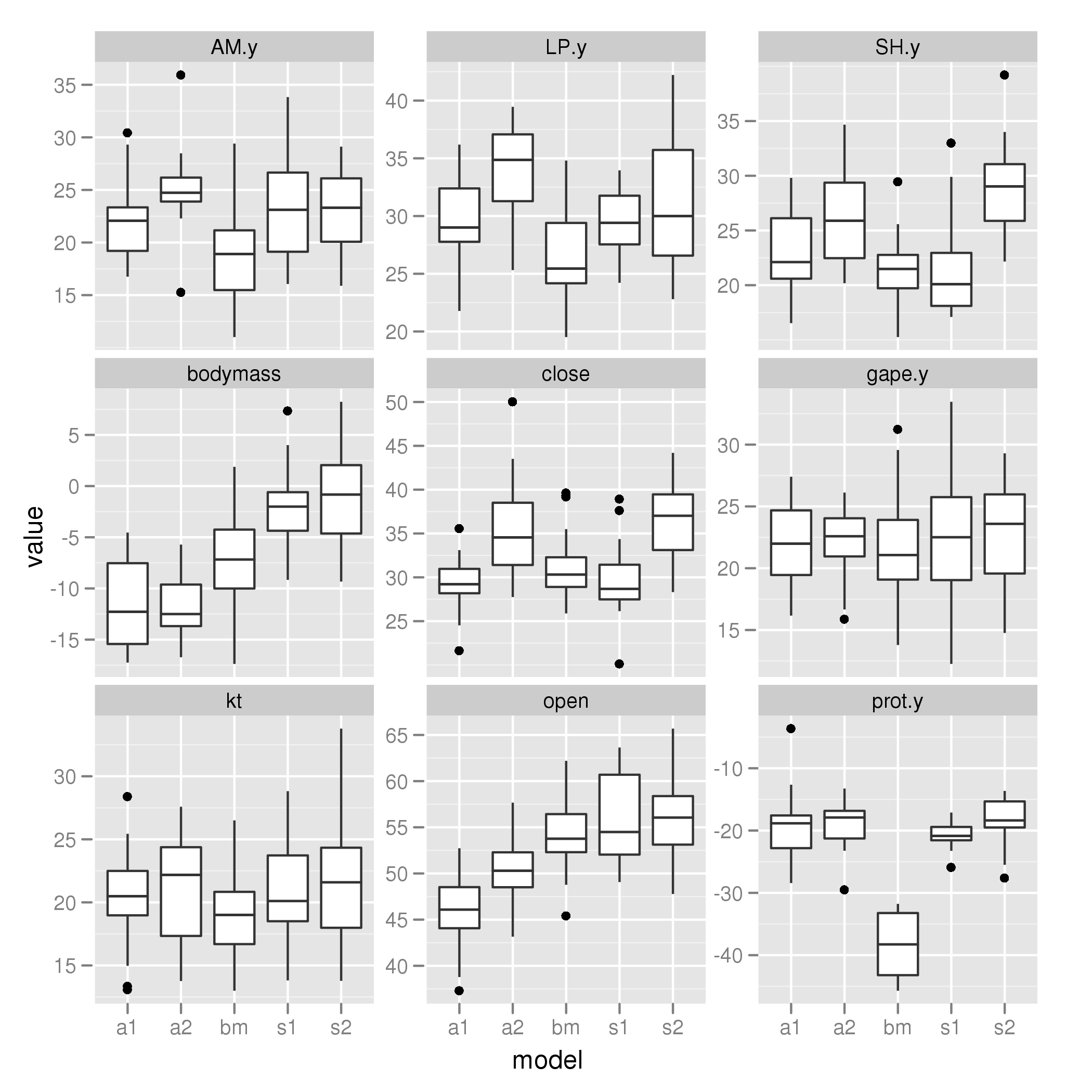
alpha
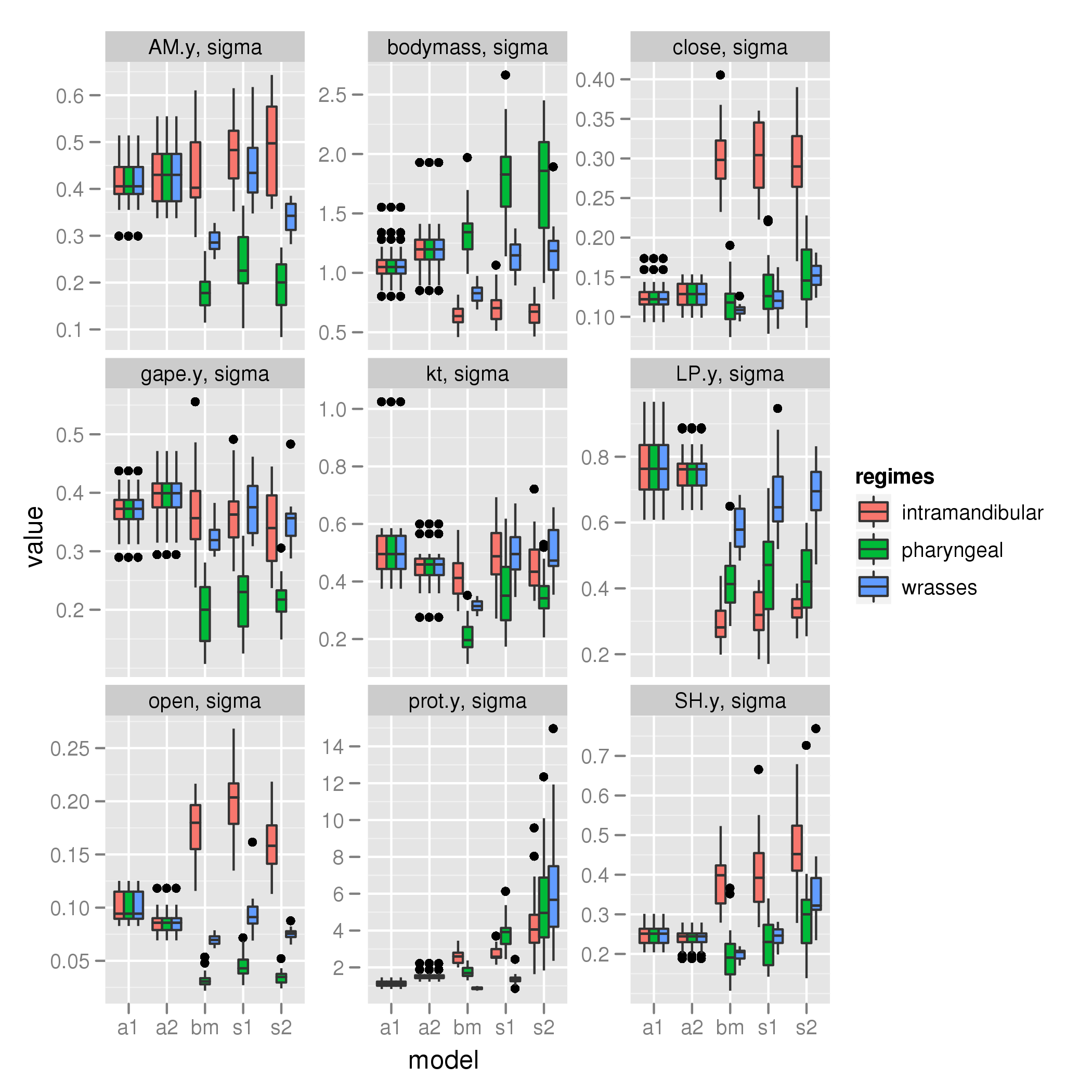
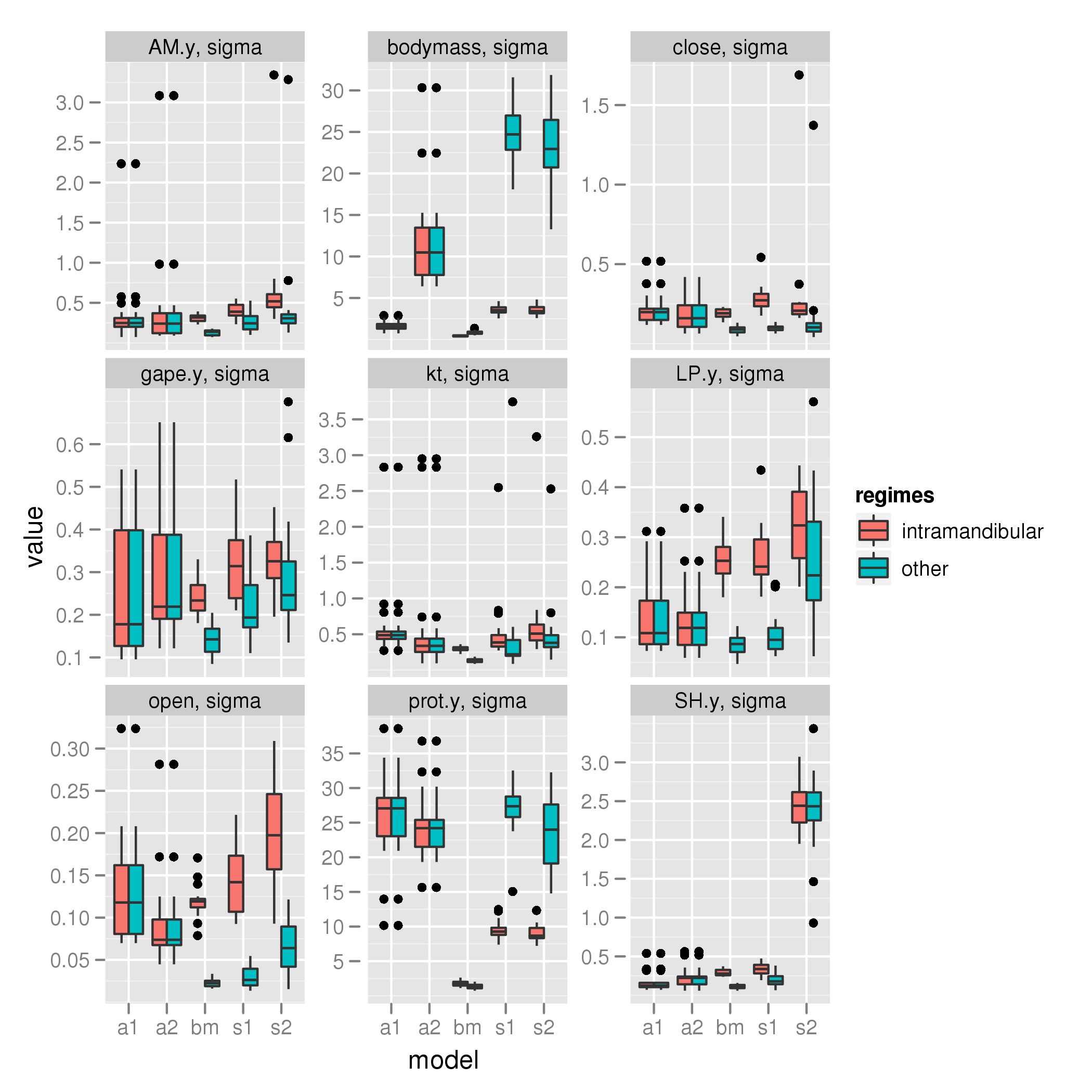
sigma
likelihood
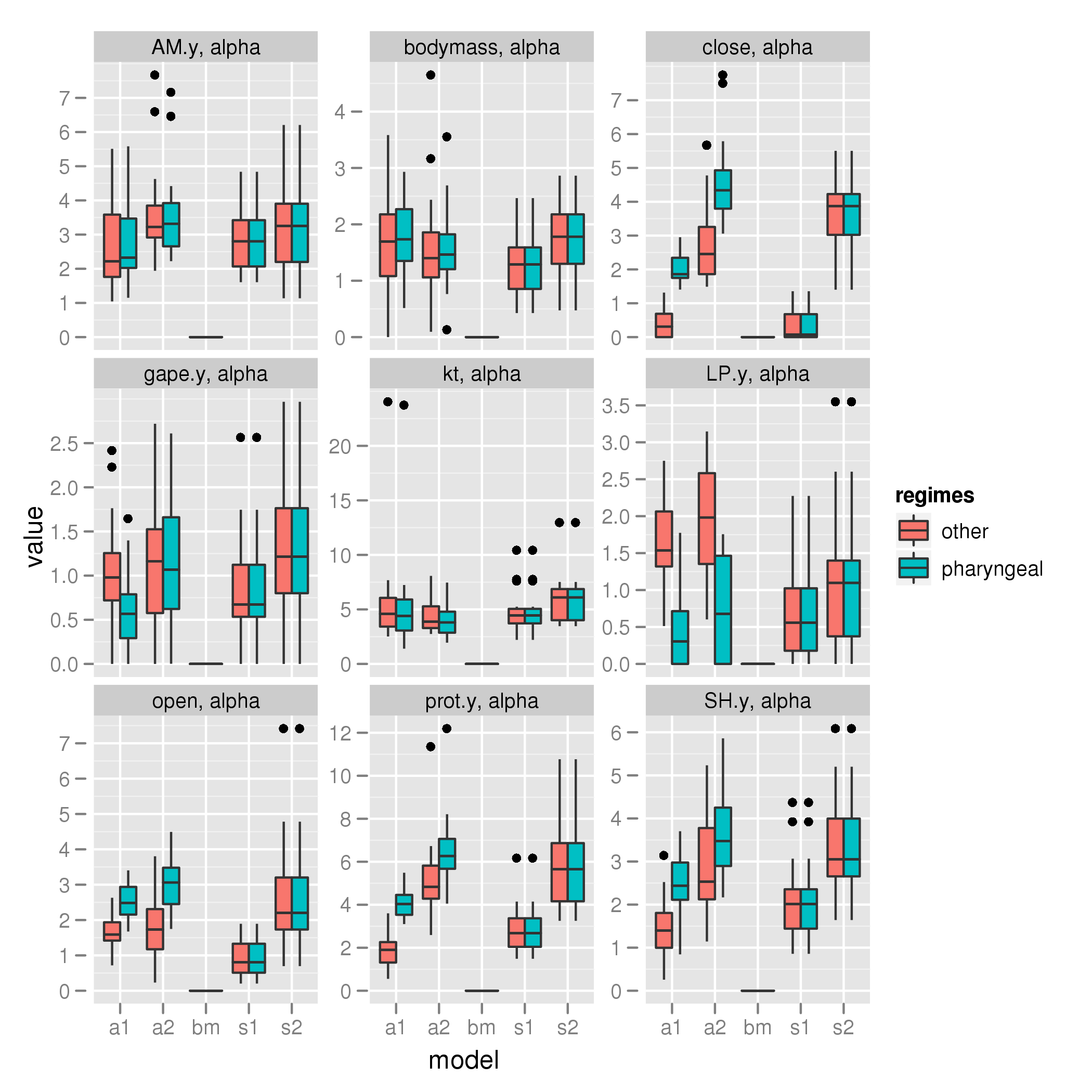
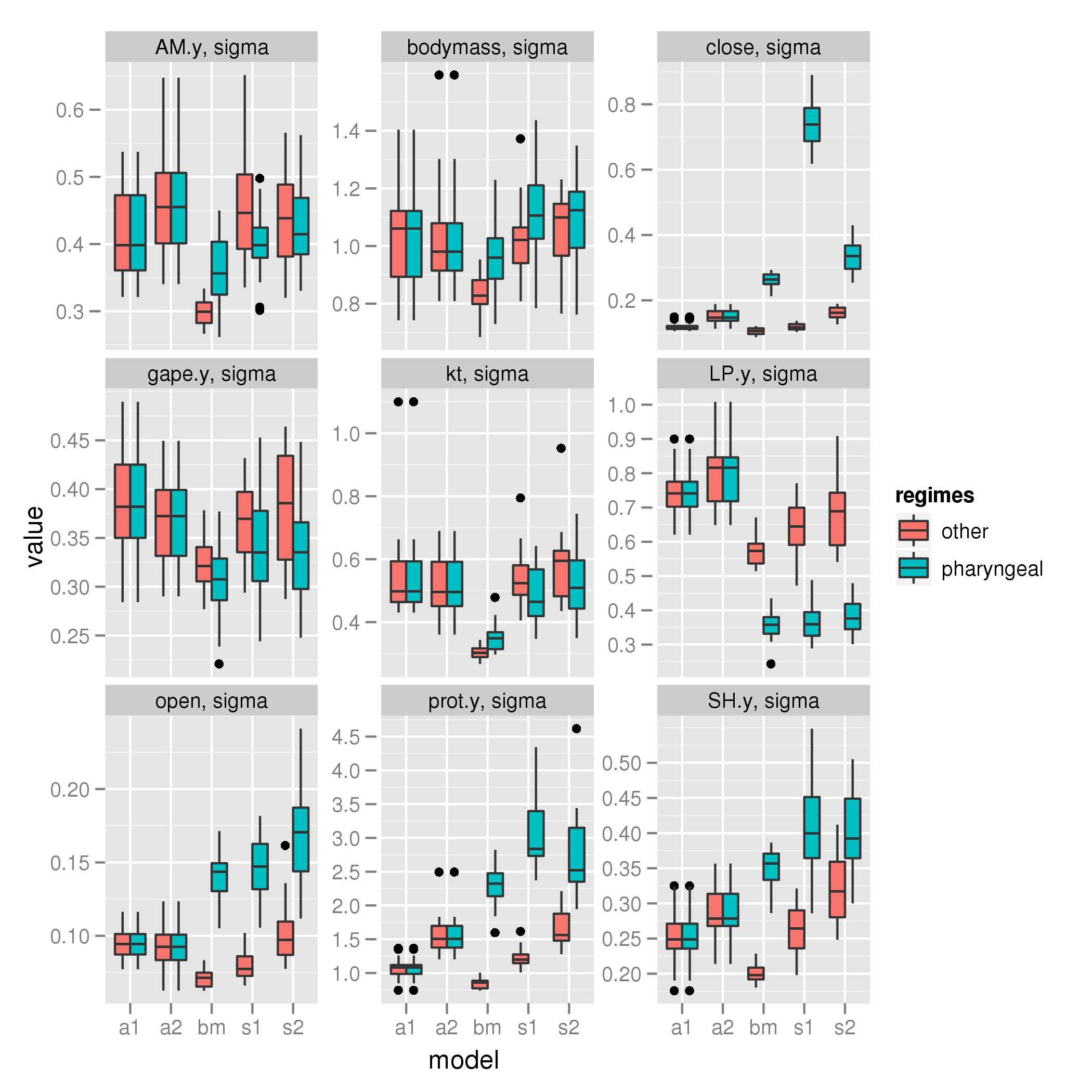
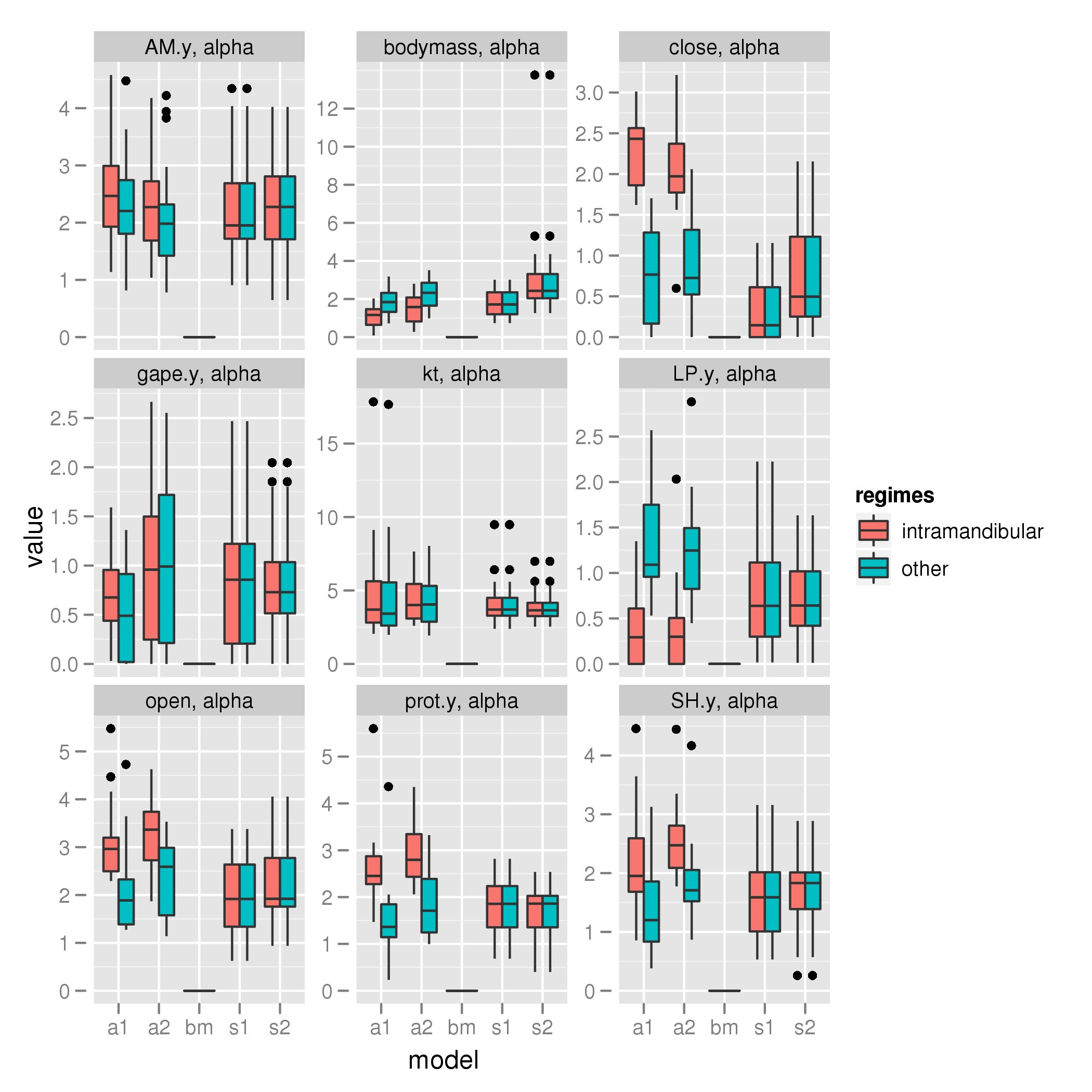
Pharyngeal shift
alpha
sigma
likelihood

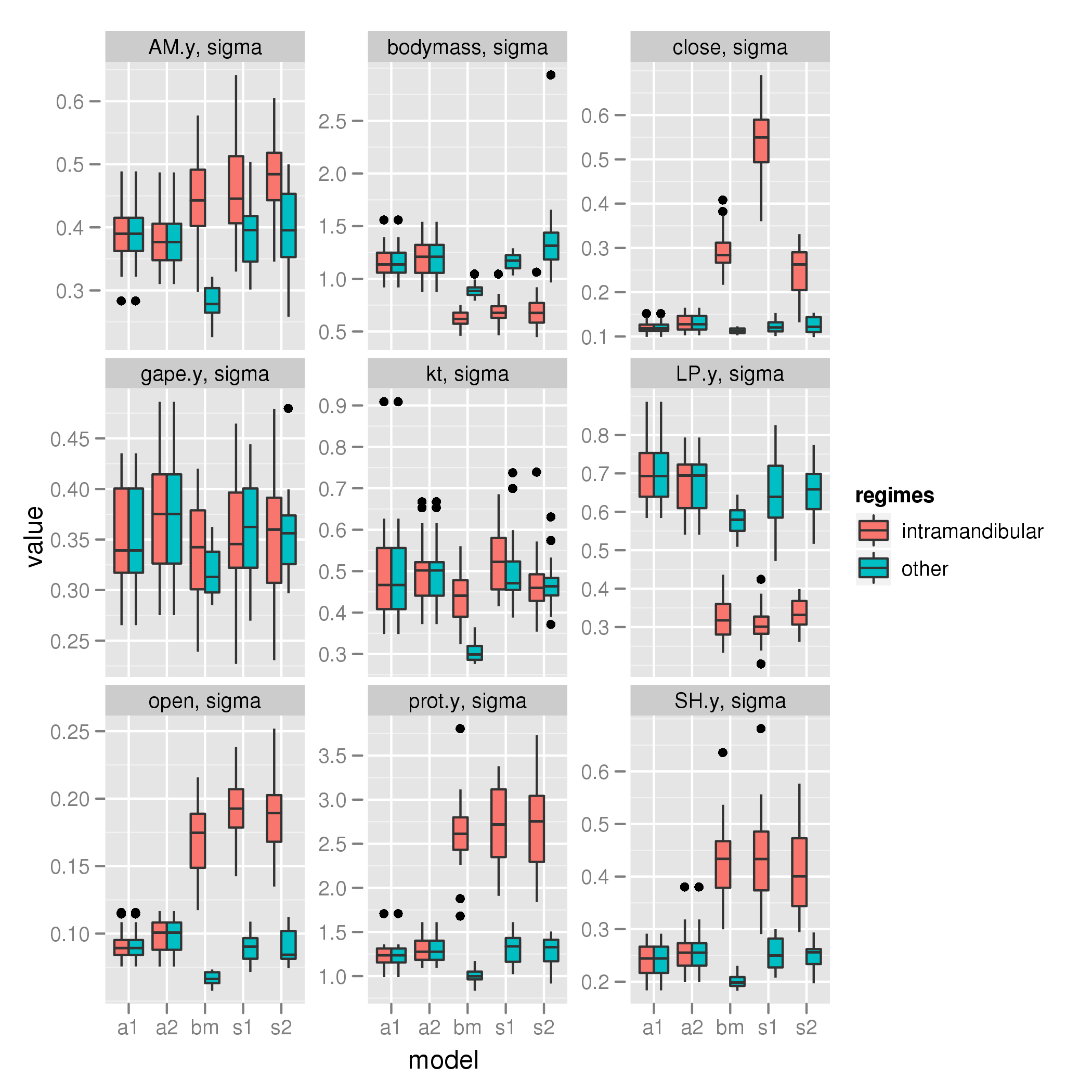
Intramandibular shift
alpha
sigma
likelihood

Intramandibular shift (parrotfish only)
alpha
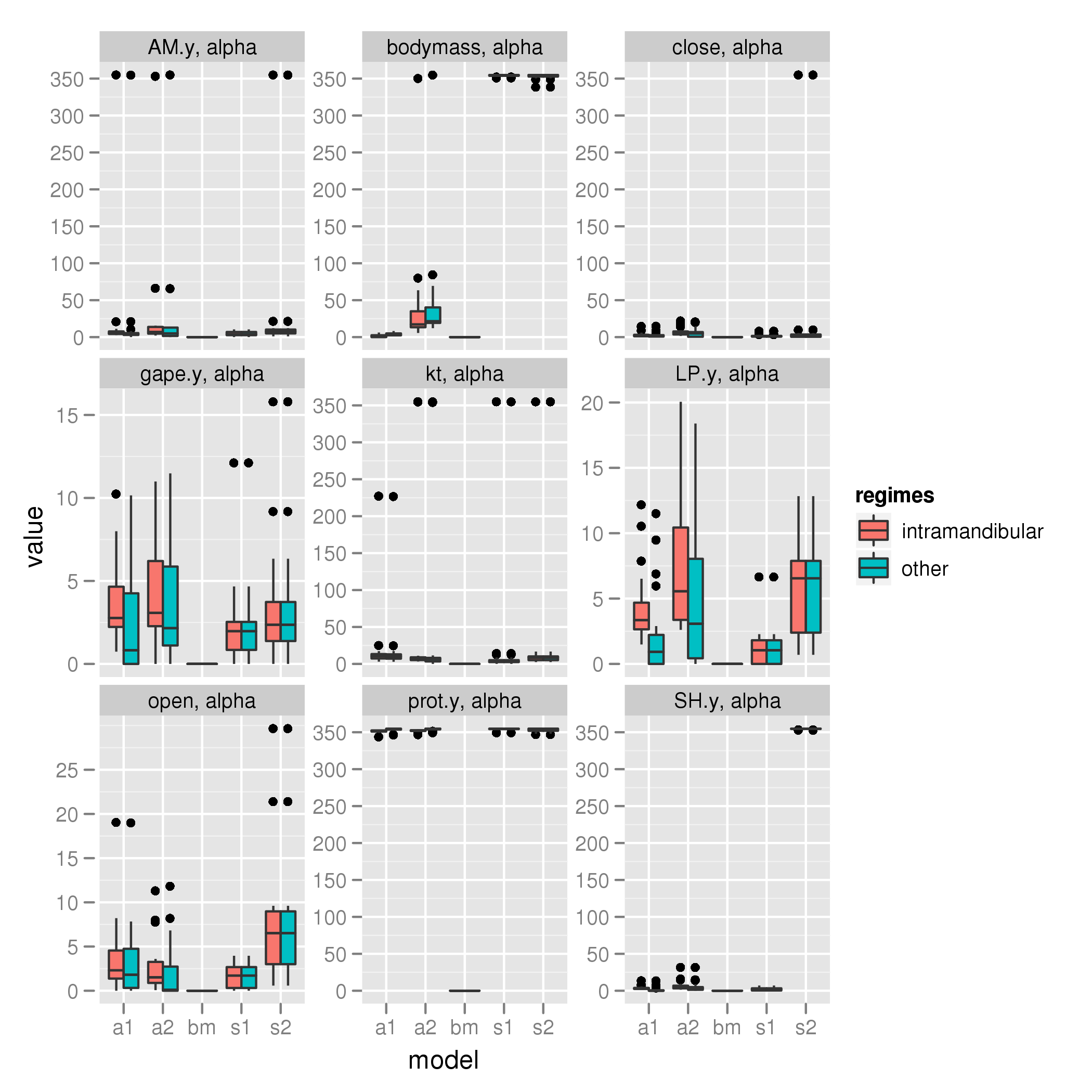
sigma
likelihood
Troubleshooting
It seems alpha is often higher in the regime that has a the higher sigma when variance is restricted to sigma. This cannot be right.
Creating unit tests:
Sigma behaves correctly in the following way. Simulate data where regimes have different sigmas.
require(wrightscape)
data(labrids)
s1_spec <- list(alpha = "global", sigma = "indep", theta = "global")
s1 <- multiTypeOU(data = dat["close"], tree = tree, regimes = pharyngeal,
model_spec = s1_spec, control = list(maxit=5000))
# Order of entries in sigma is the order regime names are given by levels (alphabetical)
names(s1$sigma) <- levels(pharyngeal)
s1$sigma["other"] <- .01
s1$sigma["pharyngeal"] <- 10
testcase <- simulate(s1)[[1]]
testcase[pharyngeal=="other" & !is.na(testcase) & testcase != 0 ] -> lowvar
testcase[pharyngeal!="other" & !is.na(testcase) & testcase != 0 ] -> highvarResults check out.
> var(lowvar)
[1] 0.009888678
> var(highvar)
[1] 12.22513
> # and the recovered estimates are pretty close to the underlying simulation parameters
> dummy <- update(s1, testcase)
> dummy$sigma
[1] 0.1015016 7.6793429
> levels(pharyngeal)
[1] "other" "pharyngeal"lowvar indeed has the lower variance (the regime with smaller sigma). And the higher sigma is estimated for the pharyngeal regime (with some expected error: 7.7 instead of 10, and 0.1 instead of 0.01).
BUT repeat with alpha:
data(labrids)
a1_spec <- list(alpha = "indep", sigma = "global", theta = "global")
a1 <- multiTypeOU(data = dat["close"], tree = tree, regimes = pharyngeal,
model_spec = a1_spec, control = list(maxit=5000))
names(a1$alpha) <- levels(pharyngeal)
data(labrids)
a1_spec <- list(alpha = "indep", sigma = "global", theta = "global")
a1 <- multiTypeOU(data = dat["close"], tree = tree, regimes = pharyngeal,
model_spec = a1_spec, control = list(maxit=5000))
names(a1$alpha) <- levels(pharyngeal)
a1$alpha["other"] <- 10
a1$alpha["pharyngeal"] <- 1e-12
a1$sigma <- c(5,5)
dat[["simulated_a1"]] <-simulate(a1)[[1]]
dummy <- update(a1, dat[["simulated_a1"]])
testcase <- dat[["simulated_a1"]]
testcase[pharyngeal=="other" & !is.na(testcase) & testcase != 0 ] -> lowvar
testcase[pharyngeal!="other" & !is.na(testcase) & testcase != 0 ] -> highvarand we get
> var(lowvar)
[1] 1.187643
> var(highvar)
[1] 0.000385144
> dummy$alpha
[1] 10.0551960 0.2670818And the variances are backwards. Note that the parameter estimates reasonably reflect the simulated regimes (other = 10.1, vs 10, intramandibular = 0.27 vs 1e-12).
How is this possible? The indexing routine is handling the sigma case correctly, and the same code is used to handle the indexing of each.
Does this have anything to do with the simulation function: No. We can recreate the issue by drawing data as iid variables instead of using the simulation along the tree:
require(wrightscape)
data(labrids)
spec = list(alpha="indep", sigma="global", theta="global")
#spec = list(alpha="global", sigma="indep", theta="global")
testcase <- dat[["prot.y"]]
other.prot.data <- testcase[intramandibular=="other" & !is.na(testcase)]
intra.prot.data <- testcase[intramandibular!="other" & !is.na(testcase)]
## Set the distribution of "other"
testcase[intramandibular=="other" & !is.na(testcase)] <- rnorm(length(other.prot.data), sd=.1)
#Set the distribution of the focal trait
testcase[intramandibular!="other" & !is.na(testcase)] <- rnorm(length(intra.prot.data), sd=10)
m <- multiTypeOU(data=testcase, tree=tree, regimes=intramandibular, model_spec=spec)
names(m$sigma) <- levels(intramandibular)
names(m$alpha) <- levels(intramandibular)The higher alpha is estimated on the regime that has higher variance
> print(m$alpha)
intramandibular other
13.121679 5.981398 The data has higher variance among the intramandibular traits. (same is true for pharyngeal). This problem does not face the sigmas model, hence appears to do with the indexing of alphas somehow.
Trying direct debug:
require(wrightscape)
data(labrids)
spec = list(alpha="indep", sigma="global", theta="global")
testcase <- dat[["prot.y"]]
other.prot.data <- testcase[intramandibular=="other" & !is.na(testcase)]
intra.prot.data <- testcase[intramandibular!="other" & !is.na(testcase)]
testcase[intramandibular=="other" & !is.na(testcase)] <- rnorm(length(other.prot.data), sd=.1)
testcase[intramandibular!="other" & !is.na(testcase)] <- rnorm(length(intra.prot.data), sd=10)
# Load internal functions
require(devtools)
load_all("..")
data <- testcase
lca <- lca_calc(tree)
multiOU_lik_lca(data, tree, intramandibular, alpha=c(2,7), sigma=c(16,5), theta=c(0.5, 0.5), Xo=0.5, lca)Looks to be keeping the alpha with sigma at least.
Check that in global ou mode, smaller alphas fit high variance better: (yup)
# Test of single OU
test <- dat[["prot.y"]]
test[!is.na(test)] <- rnorm(sum(!is.na(test)), sd=10)
ou <- list(alpha = "global", sigma = "global", theta = "global")
m1 <- multiTypeOU(data=test, tree=tree, regimes=intramandibular, model_spec=ou)
test[!is.na(test)] <- rnorm(sum(!is.na(test)), sd=.1)
m2 <- multiTypeOU(data=test, tree=tree, regimes=intramandibular, model_spec=ou)
m1$alpha # larger variance has smaller alpha
m2$alphaAll okay. But compare fits
testcase <- dat[["prot.y"]]
other.prot.data <- testcase[intramandibular=="other" & !is.na(testcase)]
intra.prot.data <- testcase[intramandibular!="other" & !is.na(testcase)]
testcase[intramandibular=="other" & !is.na(testcase)] <- rnorm(length(other.prot.data), sd=.1)
testcase[intramandibular!="other" & !is.na(testcase)] <- rnorm(length(intra.prot.data), sd=10)
# Load internal functions
require(devtools)
load_all("..")
data <- testcase
lca <- lca_calc(tree)
# expect higher alpha on "other" (alpha2) to be best, higher on alpha1 is worst
# BUT NO!
> multiOU_lik_lca(testcase, tree, intramandibular, alpha=c(2,2), sigma=c(5,5), theta=c(0.5, 0.5), Xo=0.5, lca)
[1] -1802.169
> multiOU_lik_lca(testcase, tree, intramandibular, alpha=c(5,2), sigma=c(5,5), theta=c(0.5, 0.5), Xo=0.5, lca)
[1] -248.6667
> multiOU_lik_lca(testcase, tree, intramandibular, alpha=c(2,5), sigma=c(5,5), theta=c(0.5, 0.5), Xo=0.5, lca)
[1] -110821.8
> a1_spec <- list(alpha = "indep", sigma = "global", theta = "global")
> a1 <- multiTypeOU(data=testcase, tree=tree, regimes=intramandibular, model_spec=a1_spec)
> a1$alpha
[1] 14.165247 6.579535[flickr-gallery mode=“search” tags=“phylogenetics” min_upload_date=“2011-11-28 10:15:00” max_upload_date=“2011-11-28 10:30:20”]