laptop: ubuntu upgrades: 10.04 LTS -> 10.10 -> 11.04, geesh.
Set up for running primates.R in likelihood mode with abstracted parallelization:
Carver trouble getting wrightscape installed: even after module load gsl, cannot find gsl library. Testing on zero in mpi mode, 16. Testing on farm, mpi mode with snow, 16 cores.
hmm, high (9%) memory usage on primates.R on zero… monitoring situation closely… Only .3% on farm so far, which has 24G instead of zero’s 32Gig RAM..
farm runs at 16 cores, bm vs ouch: successful. Little difference in differing optima between new and old world monkeys vs brownian motion:
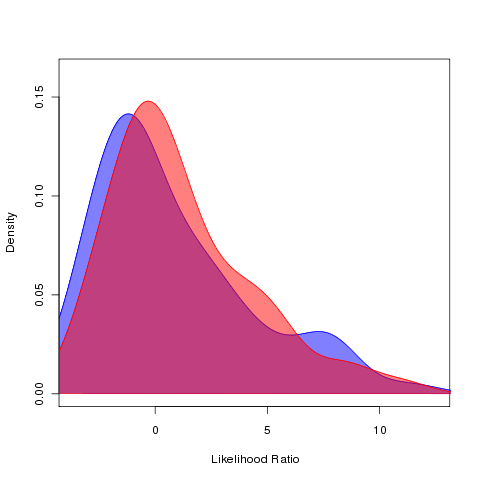
Running bm vs alphas models on primates, 64 cpu on farm
Running sigmas vs alphas model as primates2.R, 64 cpu on farm
estimate of sigmas model on primates data is very slow relative to alpha, etc. hmm..
- Running ou vs alphas on parrotfish.R 64 cpu on farm
(looks like sigmas v alphas but note sigmas is defined as all global, i.e. 1-peak ou, in the model.spec). Add labrid data to git tracking to facilitate this. Dramatically better log likelihood on the alphas model, easily supported by the bootstrap: (trait is size-corrected protrusion).
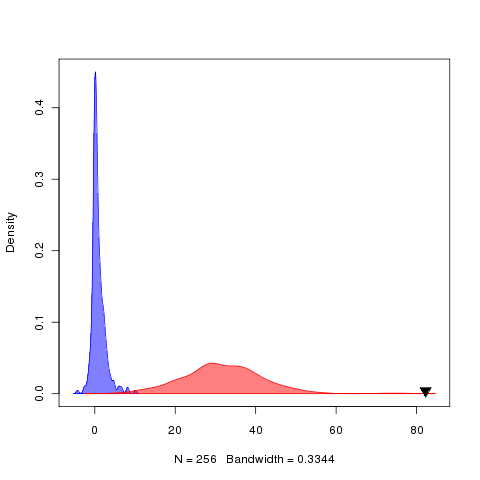
plotting for parameter distributions is supported from wrightscape, to handle the regimes grouping on matching plots. A more basic/general one is found in pmc pots. Should add a decent general plotting of parameters to mcmcTools, as well as ROC curve tools.
Updated socialR a bit to allow more minimal details specified in upload, and to work better with reporting from head node following a cluster run.