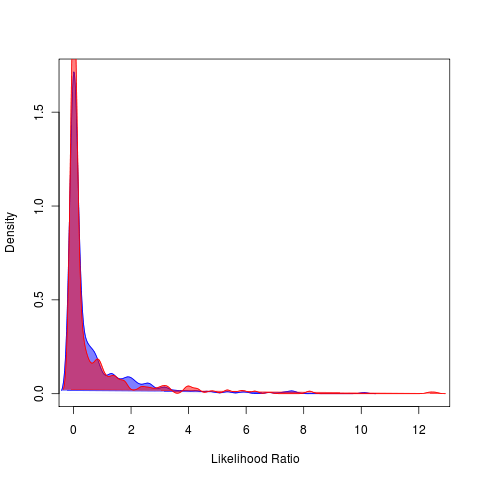
Laser simulations: First ensuring example has expected behaviour, can in general get the expected likelihood differences between fits of Yule and birth-death models.
Estimated models had been appearing identical in the runs (on left), seems to have been a bug. Distributions appear roughly the same across different tree sizes and values of death rate. General pattern of likelihood tests is reminiscent of the lambda models: high variability in likelihood ratios under bd simulations relative to the pure-birth simulations. Needs further exploration

looking at parameter estimation
(a la lambda estimation)
Left: r = b-d, should be easy to estimate. Surprised by tendancy to overestimate: r should be near 1-0.5=.5. Right, a = d/b, should be harder to estimate, but uncertainty seems small. However, note it greatly underestimates the true value of a=0.5 .
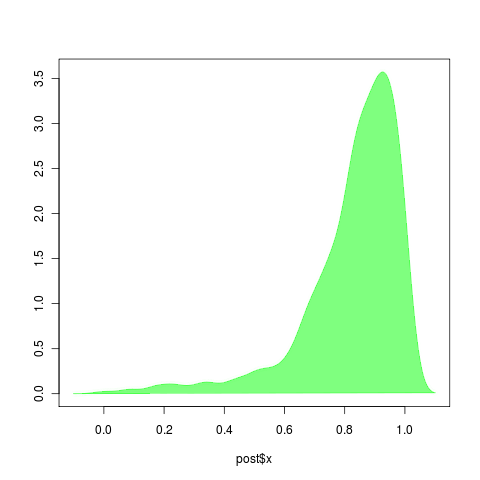
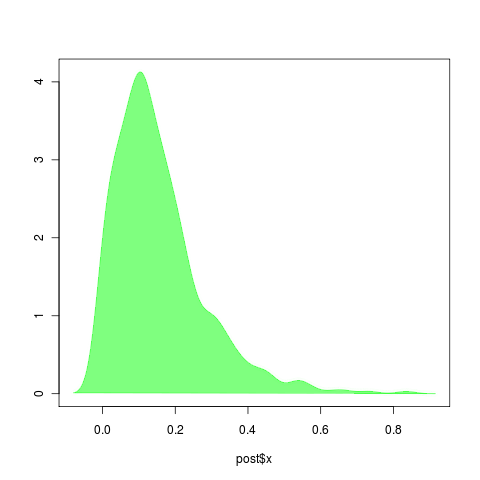
Estimates of d tend to be high (left), though estimates of birth seem reasonable? (right)
[flickr size=“small” float=“left”]https://www.flickr.com/photos/cboettig/5431673885/[/flickr]
[flickr size=“small” float=“left”]https://www.flickr.com/photos/cboettig/5431671921/[/flickr]
Pure-birth model estimates seem reasonable.
[flickr size=“small”]https://www.flickr.com/photos/cboettig/5432281984/[/flickr]
Code progress
Added parameter plotting routine (currently in demo code) and functions for handling of parameter bootstrap plotting.
To Do
the loglik function should have the option to recalculate likelihood, not just lookup the value attached to the fit object. The likelihood functions are rather buried in laser though.
For some reason bd.taxa.sim doesn’t behave properly on wiglaf.
diversitree, brownie.light, error estimates over trees vs over bootstrapping.