- Wrote the proper record structure into C code to handle reporting to R, including ensemble data.
- Wrote the R interface to the dynamics.
- Needs exploration.
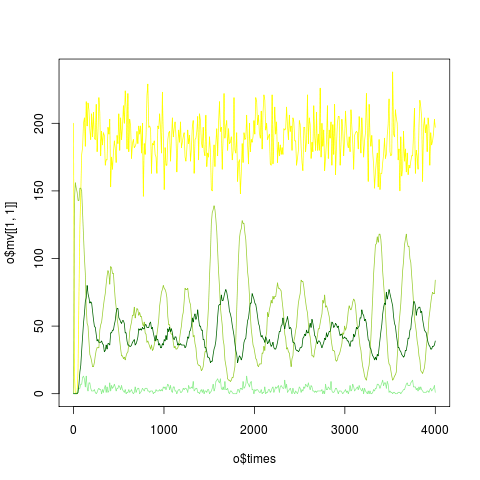
Version-stable code for parameters. Run using defaults, graph as:
png("ibm_gamma.png")
o <- gamma_beetles_ibm()
plot(o$times, o$mv[[1,1]], type='l', col="yellow")
lines(o$times, o$mv[[1,2]], col="yellowgreen")
lines(o$times, o$mv[[1,3]], col="lightgreen")
lines(o$times, o$mv[[1,4]], col="darkgreen")
dev.off()
o$parameters
b ue ul up ua ae al ap cle cap
5.000 0.000 0.001 0.000 0.010 1.300 0.100 1.500 0.200 0.100
cae V
5.000 100.000- Simulation reports actual parameters above, chosen to match the theory results from last week (linear noise approximation).
- Comparisons using ensemble data against theory coming soon.
- Single replicate .c based IBM is currently faster than the ode system (with variance dynamics).
- Note: Entry and graph above actually added on June 3rd, parameters weren’t named correctly on June 1st, so simulation wasn’t oscillating.