- Parallelized LR_bootstrap_all() and model_bootstrap_all() bootstrapping functions. Lets the bootstrapping of different models (or LRs between models) to be performed on different machines.
- Using the snowfall package in R for easy parallel computing. The parallel loop is done through the sfLapply() function.
- Had to get libraries and functions exported to all loops using sfLibrary() and sfExportAll(). R-sig-hpc mailing list was very helpful at getting this up and running at 4am.
References for parallel R
Research ——–
- Still exploring model comparisons. Trying labrid data with up to 5 partitions.
- Attempted partitioning on the Anoles data, but for some reason the ape ace() fn for discrete ancestral state reconstruction fails on this data (seems to get stuck in an infinite loop). Not obvious why this should occur; geiger’s fitDiscrete() works fine on the partitioned data. Well, was planning on rewriting the ancestral state reconstruction in simmap style anyway…
Anoles
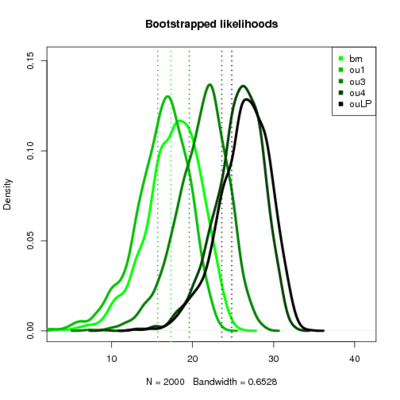
Labrids
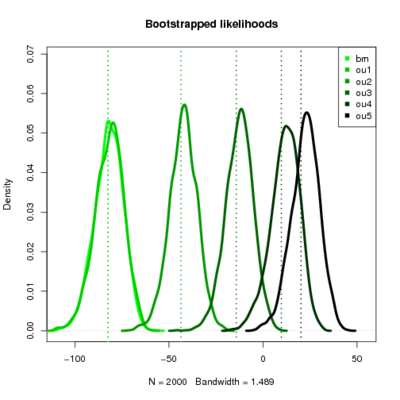
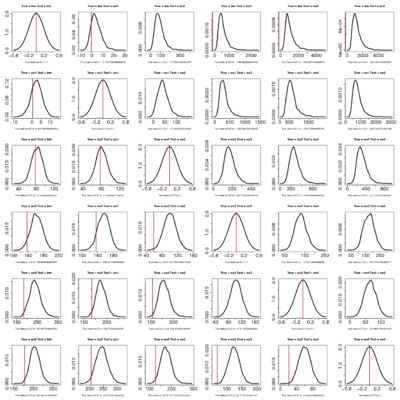
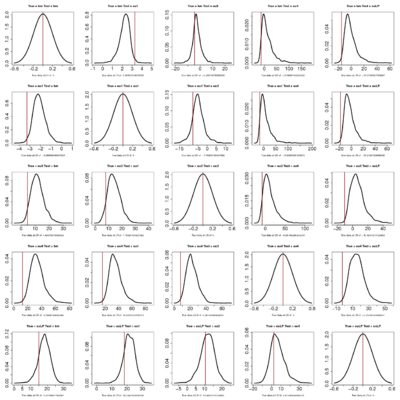
- Seems like the more complicated models continually perform better. However, the observed likelihood ratios become less and less likely themselves, even against simple models.
- Recall I simulate the dataset under the model labeled “true” and then evaluate the likelihood ratio of that model vs the test model. So along a row data is produced by simulation of the same model. The pattern is basically that the upper triangle of the matrix (simulate simple, compare to complex) always has negative LR for the true data but a very positive distribution of LRs. It’s still a bit surprising to me that when data simulated under the simpler model that the simple model should always drastically outperform the complex one in LR, but it does.
Misc Notes
Listened to some talks while coding yesterday. Listing here in case I want to return to them.
- Richard Florida What’s your city? (excellent)
- Tony Schwartz Managing your time managing your energy
- David Weinberger Everything is Miscellaneous