Last day of lectures: Lineage Diversification
Brian Moore: Rates of Lineage Diversification
- Detecting significant diversification rate variation across tree
- locate shifts along branches
- and through time (niche-filling, density dep models?)
- correlates of differential rates
- parameter estimation speciation/extinction

correlates of differential rates
- Apply Felsenstein pruning algorithm to calc likelihood and hence fit model this is just the definition of a joint probability!
- Use this to estimate the model, then do rejection sampling to create the tree painting. as in Nielsen & Yang 2002
This is applied to differential rates question in Ree et. al. 2005
- Simply ask if group A or B observed more branching events along the stochastic map. Compare to a null that randomizes the branch lengths as waiting times under pure yule model.
- Methinks this creates null trees that violate the coalescent pattern of longer branches near the base, but guess Yule is the opposite of coalescent anyway…
- Comparison method: Moore’s cross-validation approach: split tree into two parts, generate background rate from one, use that rate to create a distribution, of possible outcomes and compare that distribution to the observed rate in the test clade. Also handle uncertainty in the time of the rate transition, (Bayesian). Good when traits fall out in clades.
- Methinks should compare the two-rate directly to the single rate fitted to the entire tree, for completeness.
- Wonder where else we should start trying cross-validation approaches in comparative methods. Perhaps in comparison to the OU(LP) Anoles painting approach?
- Trickle-down problem – one rapid clade can inflate the score farther down the tree.
- AIC comparison of assigning different shift points.
r88s
Parameter estimation Bayesian approach to explore shifts in diversification rate via DPP!
### Community Phylogenetics Sharon Strauss, Jean Burns
- Community assembly: Niche, History, or random?
- (Janzen Connell hypothesis of diversity optimal radius in seed-shadow for recruitment)
- build a tree of a regional species pool
- build a tree of local community
- generate null comunities
- Methinks Not robust to incomplete sampling of the community phylogenetics null model vs comparative trait methods
Null distribution should be generated by the non-interacting model!!
Phylocom tutorial
Thoughts & Ideas scratch pad
- Density dependent estimates
Levins model (logistic) style: 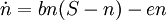
calculate probability n conditional on non-extinction for remaining time. let’s start by assuming stationarity across whole tree. Then the number of lineages observed at time t should decrease exponentially. This pattern looks just like the pure birth model or the constant rates model. When this pattern ceases to hold, we’re out of the realm of constant rates and we head into the dynamic model. What should be the signal? slower than exponential fall-off.
ha, we often look for the other pattern – accelerating rates.
- Community phylogenetics should generate nulls under evolutionary models, not random reshuffling.
(Will develop this further. Also, evening session to focus on Labrid analysis, general research progress).